Poincaré maps for analyzing complex hierarchies in single-cell data
Abstract:
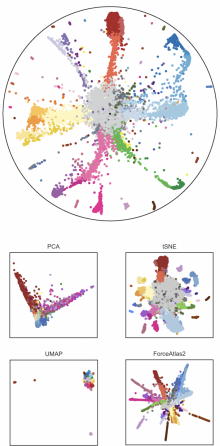 The need to understand cell developmental processes has spawned a plethora of computational methods for discovering
hierarchies from scRNAseq data. However, existing techniques are based on Euclidean geometry which is not an optimal
choice for modeling complex cell trajectories with multiple branches. To overcome this fundamental representation issue we
propose Poincaré maps, a method harnessing the power of hyperbolic geometry into the realm of single-cell data analysis.
Often understood as a continuous extension of trees, hyperbolic geometry enables the embedding of complex hierarchical data
in as few as two dimensions and well-preserves distances between points in the hierarchy. This enables direct exploratory
analysis and the use of our embeddings in a wide variety of downstream data analysis tasks, such as visualization, clustering,
lineage detection and pseudotime inference. In contrast to existing methods – which are not able to cover all those important
aspects in a single embedding – we show that Poincaré maps produce state-of-the-art two-dimensional representations of
cell trajectories on multiple scRNAseq datasets. Specifically, we demonstrate that Poincaré maps allow in a straightforward
manner to formulate new hypotheses about biological processes which were not visible with the methods introduced before.
Moreover, we show that our embeddings can be used to learn predictive models that estimate gene expressions of unseen cell
populations in intermediate developmental stages.
The need to understand cell developmental processes has spawned a plethora of computational methods for discovering
hierarchies from scRNAseq data. However, existing techniques are based on Euclidean geometry which is not an optimal
choice for modeling complex cell trajectories with multiple branches. To overcome this fundamental representation issue we
propose Poincaré maps, a method harnessing the power of hyperbolic geometry into the realm of single-cell data analysis.
Often understood as a continuous extension of trees, hyperbolic geometry enables the embedding of complex hierarchical data
in as few as two dimensions and well-preserves distances between points in the hierarchy. This enables direct exploratory
analysis and the use of our embeddings in a wide variety of downstream data analysis tasks, such as visualization, clustering,
lineage detection and pseudotime inference. In contrast to existing methods – which are not able to cover all those important
aspects in a single embedding – we show that Poincaré maps produce state-of-the-art two-dimensional representations of
cell trajectories on multiple scRNAseq datasets. Specifically, we demonstrate that Poincaré maps allow in a straightforward
manner to formulate new hypotheses about biological processes which were not visible with the methods introduced before.
Moreover, we show that our embeddings can be used to learn predictive models that estimate gene expressions of unseen cell
populations in intermediate developmental stages.
https://www.nature.com/articles/s41467-020-16822-4
tr-poincare-2019.djvu
tr-poincare-2019.pdf
tr-poincare-2019.ps.gz
@article{klimovskaia-2020,
author = {Klimovskaia, Anna and Lopez-Paz, David and Bottou, L{\'e}on and Nickel, Maximilian},
isbn = {2041-1723},
journal = {Nature Communications},
number = {1},
pages = {2966},
title = {Poincar\'{e} maps for analyzing complex hierarchies in single-cell data},
ty = {JOUR},
volume = {11},
year = {2020},
url = {http://leon.bottou.org/papers/klimovskaia-2020},
}